The Lilley Group, and collaborators, have published a simple and cost-effective method to capture the RNA-binding proteome in Nature Biotechnology.
To date, the major method used to determine the repertoire of RNA-binding proteins in cells has been limited to the study of proteins that bind polyadenylated-RNA since it uses oligo(dT) to purify RNA:protein adducts. It is thus not suitable for the study of proteins that bind non-coding RNAs, including lincRNAs, nor is it suitable for use in species where there is short and/or infrequent mRNA polyadenylation, such as in many microorganisms. In collaboration with researchers at the MRC Toxicology Unit, the MRC LMB and the University of York, the Lilley Group have developed a new method, dubbed OOPS (orthogonal organic phase separation), to study the RNA-binding proteome. OOPS is not only able to enrich RNA-bound proteins involving the majority of cellular RNA species, irrespective of whether they are poly-adenylated, but also requires much less material than the established methods, widening its applicability.
The OOPS approach re-purposes acid guanidinium thiocyanate-phenol-chloroform (AGPC) extraction, commonly referred to as the 'TRIzol protocol' after the branded reagent, which has been used for decades to enrich and partition RNA from other cellular components. The Trizol protocol separates RNA and protein in aqueous and organic phases respectively, and instructs the researcher to avoid the interface. However, the Lilley Group and their collaborators reasoned that RNA:protein adducts generated by UV cross-linking will be highly enriched at the interface, including rRNAs, tRNAs, mRNAs, sRNAs and other non-coding RNAs. The OOPS protocol therefore begins from this interface, with repeated phase separations to ensure removal of non-RNA-bound protein from the resulting final interface. Proteins at this final interface can then be recovered by enzymatically digesting the RNA and repeating the Trizol phase separation, taking the resulting protein-rich organic phase.
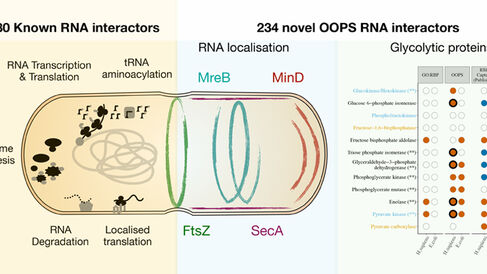
To demonstrate the versatility of the OOPS method, the authors applied OOPS to catalogue the RNA-binding proteome of three human cell lines, including the first human non-tumoral cell line, uncovering surprising roles as RNA-binding proteins for many metabolic enzymes. They also showed that the extent of interaction between RNA and protein is significantly modulated when microtubule polymerisation is inhibited. Finally they captured the first RNA-binding protein catalogue of a bacterium, using Escherichia coli as their test system, again demonstrating RNA-binding of many metabolic enzymes; a function which may be conserved through evolution. They also uncovered E. coli RNA-binding proteins with intriguing subcellular distributions, which could conceivably coordinate RNA localisation; a process that has been recently proposed to occur in microbes, but which has not been widely studied.
Amongst the contributing co-authors of this paper are 2017-2018 Part III Systems Biology student, Mie Monti, and 2017-2018 Part II Biochemistry student, Dan-Mircea Mirea.
