
This section summarizes the work going on in the lab. The links to individual people's web pages give you more information on particular areas. Our work covers a number of areas in the biochemistry and molecular biology of photosynthetic organisms, and the evolutionary processes that gave rise to them. We trace the lineage of our interest in photosynthesis to Robin Hill, one of the great pioneers of photosynthesis biochemistry, and our interest in the chloroplast genome and its evolution to Fred Sanger and his application of his DNA sequencing methodology to the mitochondrial genome.
Our present interests lie in three main complementary areas:
- the biochemistry of photosynthesis, and its exploitation for biotechnology, including renewable energy production and bioremediation of waste materials
- the organization and evolution of the chloroplast genome – including in the malaria parasite Plasmodium and their close relatives the dinoflagellate algae
- novel applications of evolutionary biology – the evolution of medieval manuscripts
We were founder members of the Algal Bioechnology Consortium, a Cambridge-based interdisciplinary group of scientists aiming to use algae for a number of different applications in bioenergy and biotechnology.
We also have close links with Martin Jones, Mim Bower, Harriet Hunt and Diane Lister of the McDonald Institute for Archaeological Research, studying a range of archaeogenetic questions. Please look at their entries on this site or the McDonald Institute for more information.

Double Nobel Laureate Fred Sanger opening the lab after its refurbishment

Photosynthesis biochemistry pioneer Robin Hill
The biochemistry of photosynthesis...
The figure below shows a schematic representation of the electron transfer chain in cyanobacteria. (These are oxygenic photosynthetic bacteria, also known as blue-green algae, and are responsible for a large part of global oceanic productivity.)

The electron transport chain in cyanobacteria. © CJH 2005
The figure shows the two photosystems (PSI and PSII) linked by the cytochrome b6f complex. Two soluble electron carriers can accept electrons from cytochrome f. These are plastocyanin, which contains copper, and cytochrome c6, which contains haem. Cytochrome c6 substitutes for plastocyanin under conditions of copper deprivation. In chloroplasts of higher plants, the situation is simpler. There is no cytochrome oxidase and, if you believe the textbooks, there is no cytochrome c6. However, in 2002, we reported that there is a cytochrome c6 in plants, which we have named cytochrome c6A. However, this protein does not function as a simple substitute for plastocyanin in photosynthetic electron transfer - its real function remains a mystery, which we are working to solve. A key to this is likely to be the presence in cytochrome c6A of an insertion containing two conserved cysteine residues (marked in yellow in the figure below).
For more information, see:
"Modulation of heme redox potential in the cytochrome c6 family" Worrall JAR, Schlarb-Ridley BG, Reda T, Marcaida MJ, Moorlen RJ, Wastl J, Hirst J, Bendall DS, Luisi BF, Howe CJ (2007) Journal of the American Chemical Society 129:9468-9475
"A new function for an old cytochrome" Molina-Heredia FP, Wastl J, Navarro JA, Bendall DS, Hervas M, Howe CJ, De La Rosa MA (2003) Nature 424:33-34

Predicted structures of plant cytochrome c6A (left) and the well-characterized cyanobacterial protein (right). Picture produced by Derek Bendall © CJH 2005.
...and its exploitation for biotechnology, including renewable energy production.
Algae and photosynthetic bacteria offer very exciting prospects for a range of biotechnological applications, including renewable energy production. We are interested in exploiting these organisms, either by simple manipulation of environmental conditions, or by genetic modification. We are particularly interested in developing the use of purple photosynthetic bacteria, such as Rhodopseudomonas palustris, for example in using it to generate hydrogen from glycerol, a waste product from biodiesel manufacture.
For more information see:
“The purification of crude glycerol derived from biodiesel manufacture and its use as a substrate by Rhodopseudomonas palustris to produce hydrogen” Pott RWM, Howe CJ, Dennis JS (2014) Bioresource Technology 152:464-470.
“Thylakoid terminal oxidases are essential for the cyanobacterium Synechocystis sp. PCC 6803 to survive rapidly changing light intensities” Lea-Smith DJ, Ross N, Zori M, Bendall DS, Dennis JS, Scott SA, Smith AG, Howe CJ. (2013) Plant Physiology 162:484-495.
Large-scale industrial cultivation of micro-organisms poses technical problems, such as contamination, and in collaboration with Alison Smith in the Department of Plant Sciences, we are looking at nutritional interactions between organisms in culture, with the aim of understanding how to create stable large-scale culture systems.
For more information see:
“Biodiesel from algae: challenges and prospects” Scott SA, Davey MP, Dennis JS, Horst I, Howe CJ, Lea-Smith DJ, Smith AG (2010) Curr. Opinion. Biotech. 21:277-286.
We have pioneered the development of "biophotovoltaic" devices, which allow direct electricity prduction through photosynthesis. These devices contain algal or cyanobacterial cells in a compartment containing an electrode, the anode. On illumination, electrons are passed from the photosynthetic electron transfer chain and thence to the anode. They then flow through a circuit to the second compartment in the device, containing the cathode. Depending on how the device is configured, it can be used primarily for current generation, or for hydrogen production in the second compartment. We can also use land plants in similar devices, and this led to us developing the ‘Moss Table (http://www.youtube.com/watch?v=Tw7JcOHNZlY) and a moss-powered radio (http://www.youtube.com/watch?v=YV8nKPdu_Xs).
These devices have been exhibited internationally, won design and other awards, and are also being developed as educational toolkits for use in schools and other public awareness activities.
For more information see:
“Hydrogen production through oxygenic photosynthesis using the cyanobacterium Synechocystis sp. PCC 6803 in a bio-photoelectrolysis cell (BPE) system.” McCormick AJ, Bombelli P, Lea-Smith DJ, Bradley RW, Scott AM, Fisher AC, Smith AG, Howe CJ. (2013) Energy & Environmental Science 6:2682-2690.
“Comparison of power output by rice (Oryza sativa) and an associated weed (Echinochloa glabrescens) in vascular plant biophotovoltaic (VP-BPV) systems” Bombelli P, Iyer DMR, Covshoff S, McCormick AJ, Yunus K, Hibberd JM, Fisher AC, Howe CJ (2013) Applied Microbiology and Biotechnology 97:429-438
“Photosynthetic biofilms in pure culture harness solar energy in a mediatorless bio-photovoltaic (BPV) cell system” McCormick AJ, Bombelli P, Scott AM, Philips A, Smith AG, Fisher AC, Howe CJ. (2011) Energy & Environmental Science 4:4699-4709

Algal cells and an example of the biophotovoltaic device. © CJH 2010.
The organization and evolution of the chloroplast genome
Chloroplasts of plants and algae contain a genome that is the remnant of the genome of the photosynthetic bacterium that gave rise to chloroplasts some 1.5 billion or more years ago. This genome is typically about 150 kbp in size and contains 120 or more genes, encoding polypeptides of the photosynthetic machinery, as well as proteins and RNAs required for their expression and many other functions. Understanding the organization and expression of the chloroplast genome is a long-standing theme in the lab.
The last major group of organisms to have their chloroplast genome characterized was the dinoflagellate algae. These ecologically important organisms are responsible for red tides (when their prolific growth turns oceans red), many instances of shellfish poisoning, and significant oil deposits. In addition, they are symbionts with coral cells, and provide the corals with nutrient. An important group of the dinoflagellate algae (which contain a light-harvesting pigment called peridinin) have a chloroplast genome completely different from other algae and plants.

The marine dinoflagellate Amphidinium carterae (size approximately ten microns). Photo by Edmund Nash © CJH 2004
Adrian Barbrook in our group showed a few years ago that the dinoflagellate Amphidinium operculatum has lost the conventional chloroplast genome. Most of the genes usually found there are believed to have moved to the nucleus. The rest are located on small plasmids or 'minicircles' of about 2.5 kbp, most of which contain a single gene - though some have two or three, and there are also 'empty' minicircles that have none! The dozen or so genes that the chloroplast has retained encode components of the light reactions of photosynthesis.
We are interested in understanding how these minicircles are maintained, where they are located, and how other proteins re-enter the chloroplast. The minicircles may be useful genetic markers for different strains of coral symbionts, and help us to understand the process of 'coral bleaching', when corals expel their dinoflagellate symbionts, with potentially catastrophic results. The discovery of this highly reduced chloroplast genome also raises questions about why genes have moved from chloroplast to nucleus during evolution, and why some have been retained in the chloroplast.
Dinoflagellate chloroplast transcripts show some unexpected modifications, such as the addition of a poly(U) tail at the 3’ end. We are interested to understand the function of this and how it has evolved. Some dinoflagellate species have relatively recently lost their chloroplasts and replaced them with new ones cannibalised from other algae. Surprisingly, these newly-acquired chloroplasts have been subject to the same polyuridylylation. Have the new chloroplasts been enslaved by this process, or are they cunningly operating ‘in stealth mode’ so the host doesn’t realise anything has changed?
A big surprise in recent years in work on chloroplast DNA was the demonstration that Plasmodium, the organism that causes malaria, has a remnant chloroplast and was once a (presumably free-living) photosynthetic protozoan. This chloroplast, which is still essential for Plasmodium to survive, represents an attractive target for antimalarials. We are interested in understanding the expression and control of the Plasmodium chloroplast genome. We have identified some of the steps involved in post-transcriptional processing, and some of the proteins that may be responsible for these.
For more information see:
“Evolution of chloroplast transcript processing in Plasmodium and its chromerid algal relatives” Dorrell RG, Drew J, Nisbet RER, Howe CJ (2014) PLoS Genetics 10:e100408.
“Functional remodeling of RNA processing in replacement chloroplasts by pathways retained from their predecessors.” Dorrell RG & Howe CJ (2012) Proc Natl Acad Sci USA. 109:18879-18884.
"The remarkable chloroplast genome of dinoflagellates" Howe CJ, Nisbet RER, Barbrook AC. (2008) Journal of Experimental Botany 59:1035-1045
Novel applications of evolutionary biology – the evolution of medieval manuscripts
We have a long-standing interest in using sequence data to answer questions in evolutionary biology, and have also been closely involved in work on novel applications of phylogenetic techniques, especially in studying the texts of manuscripts. In the days before printing, manuscripts were copied by scribes, who introduced changes - either deliberately or accidentally. For a long time, scholars have used the distribution of variations among different extant versions of a text to determine which were copied from the same earlier version and produce a stemma (plural: stemmata), a tree showing these relationships. This is known as stemmatic analysis, and was pioneered by the German scholar Karl Lachmann in the 19th century.

The scribe Jean Mielot (from 'Scribes and Illuminators', C. de Hamel, British Museum Press).
The accumulation and propagation of changes to a text during copying has clear parallels with the accumulation and propagation of mutations in DNA sequences during replication, and the construction of phylogenetic trees. We showed some time ago that computer programs used for phylogenetic analysis of sequence data could also be used very easily to analyse the texts from medieval manuscripts and produce stemmata that were consistent with the ideas of manuscript scholars.
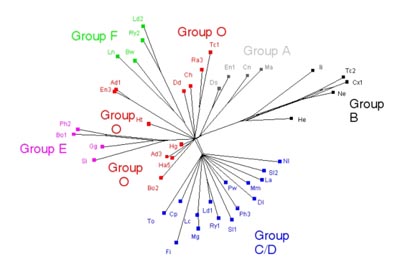
Phylogenetic analysis of the Prologue to The Wife of Bath's Tale, in Chaucer's Canterbury Tales (from Barbrook AC, Howe CJ, Blake N & Robinson P (1998) Nature 394 839). Each two- or three-letter symbol is a different extant manuscript. The groups A-F, O indicate clusters identified by conventional scholarship. © CJH 2005
We collaborate with scholars working on manuscripts ranging from the Canterbury Tales to the Greek New Testament to test and refine the use of phylogenetic programs in studying the evolution of texts. An area of particular interest is contamination, where a scribe used more than one source simultaneously when making a copy. You can see this in the picture above, where the scribe has a second book open, as well as the one he is copying from directly. This is analogous to recombination and lateral gene transfer, and we are looking at the applicability of network phylogenetic techniques and other methods for studying manuscripts that show contamination. For more information on this, see Heather Windram's page. Funding for PhD and other places in such an interdisciplinary topic is not always easy to arrange, but the application of network phylogenetic techniques to manuscripts is a very promising area, and we have many different sets of manuscripts to study.
For more information see:
“Phylomemetics - evolutionary analysis beyond the gene” Howe CJ, Windram HF. (2011) PLoS Biology 9:e1001069.
"The phylogeny of the Canterbury Tales" Barbrook AC, Howe CJ, Blake N, Robinson P (1998) Nature 394:839.
