Research
Macromolecular assemblies of transport and riboregulation
RNA-mediated regulation
In the Luisi group we are interested in understanding the mechanisms by which bacteria utilise RNA to aid development and to respond to environmental stimuli. Our research in this area can be broadly divided into two main overlapping themes as described below:
Ribonucleases in bacteria
The process of RNA turnover following transcription is vital in the regulation and quality control of gene expression. In bacteria, key components of the RNA degradation machinery are often physically associated in higher order complexes. A prime example of this is the RNA degradosome assembly found in many proteo-bacteria. We are particularly interested in studying this RNA degrading machine in Escherichia coli and other related organisms. We use a combination of structural, biophysical and biochemical techniques to understand how this complex assembles and its mechanism of action on diverse RNA substrates.
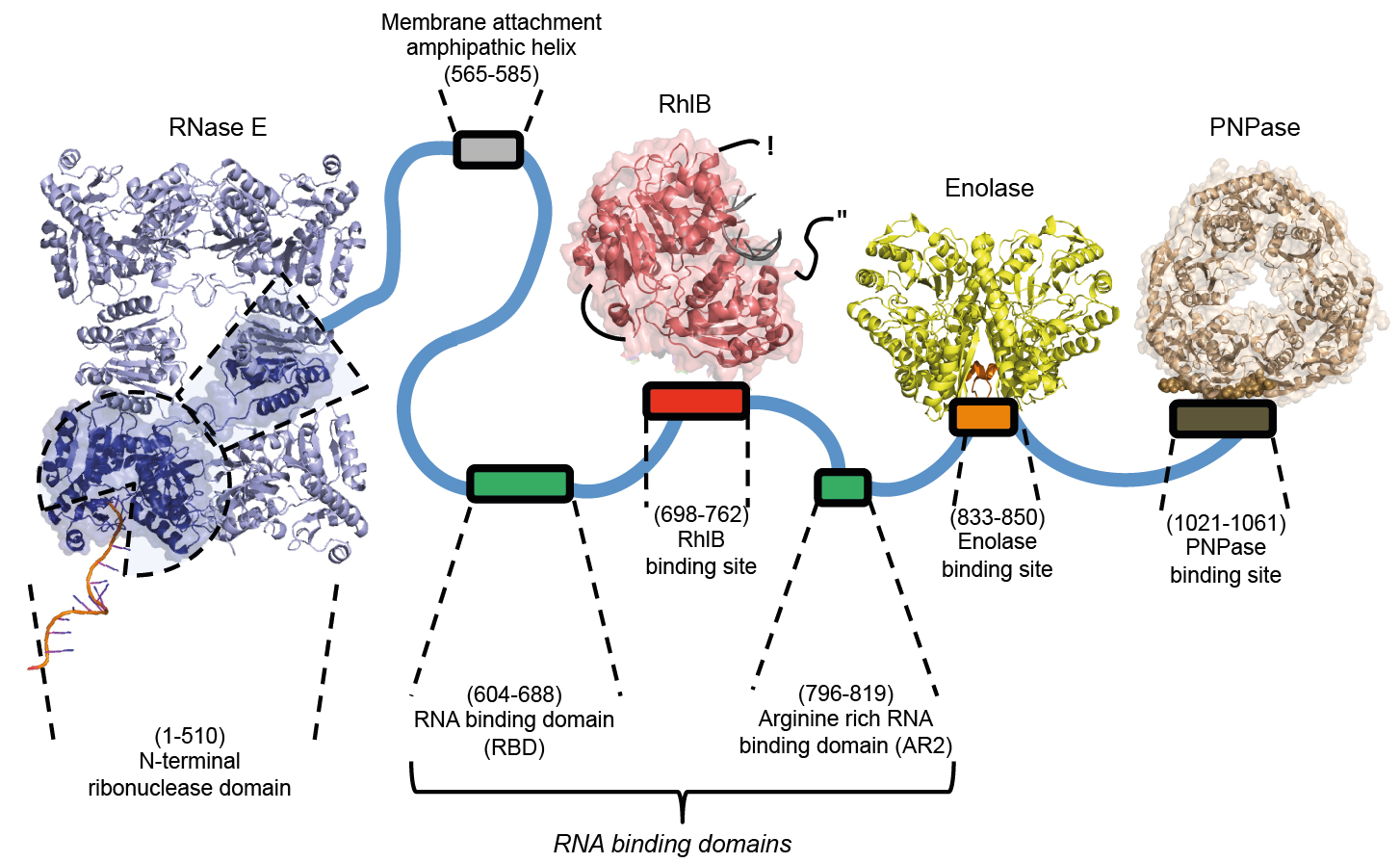
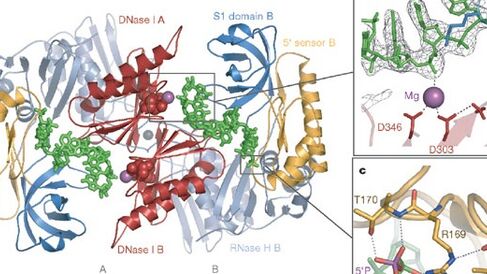
The RNA degradosome of E. coli (Hardwick and Luisi, RNA biology, 2013)
Bacterial RNA chaperone proteins
A second major area of interest of the Luisi group is the study of RNA chaperone proteins in bacteria, particularly proteins involved in assisting the action of small non-coding RNAs (sRNAs). sRNAs are versatile modulators of gene expression, and their actions are facilitated by protein chaperones with which they cooperate to achieve specificity and rapid response. In the last decade sRNAs have emerged as an important class of gene regulators in diverse bacteria that contribute to intricate post-transcriptional networks and provide controlled responses to stress, metabolic changes and signals. To date, a small number of proteins have been identified that are involved in sRNA mediated regulation of numerous bacterial species, including Hfq, CsrA and more recently ProQ like proteins. We are interested in understanding structurally how these proteins are able to recognise and engage their RNA substrates. We have been able to determine the structure of the Hfq chaperone protein in complex with the sRNA RydC (below) to reveal for the first time the detailed molecular mechanism used in this protein:RNA interaction.
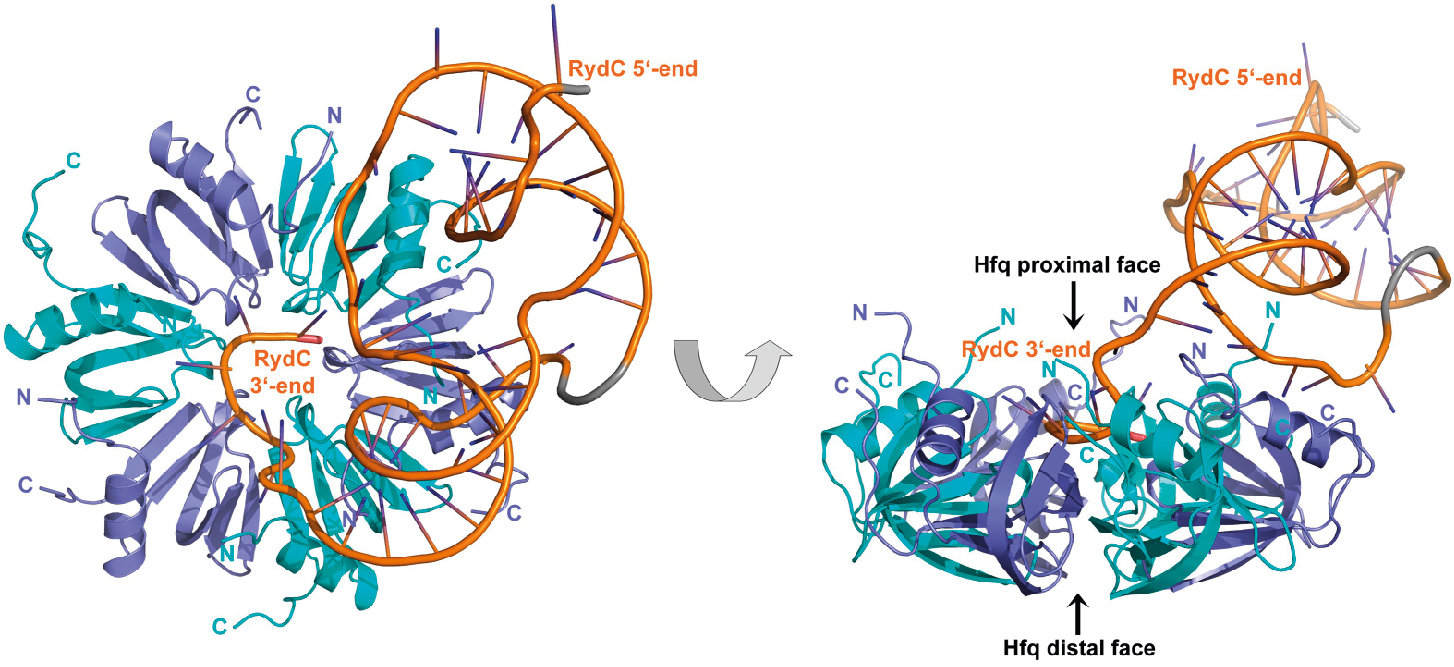
The crystal structure of Hfq bound to the sRNA RydC (Dimastrogiovanni et al, Elife, 2014)
Transport across membranes
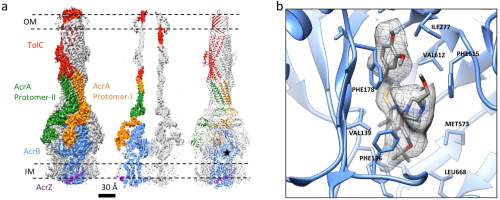
Fig 1. The RND-type tripartite multidrug efflux pump AcrABZ-TolC with inhibitor bound at 3.6 angstrom resolution
For more than two billion years, bacteria have likely encountered natural compounds of enormous chemical diversity, often synthesized by competitors, and most of which are extremely harmful. The presence of such toxic compounds and other environmental chemicals has likely sustained a tremendous selective pressure to counter the deleterious effects, and bacteria have met this challenge through various resistance mechanisms. With the clinical application of antibiotics over the last 70 years, which often resemble the compounds encountered during bacterial evolution, selective pressures have driven bacteria to use their well-developed protective mechanisms to evade these antimicrobial drugs. One common mechanism by which bacteria achieve drug resistance is through the activity of transporters that drive the efflux of toxic compounds from the cell. To date, six families of transporters have been identified that contribute to multidrug resistance in bacteria, and these can be grouped into primary-active transporters, which use the energy of ATP binding and hydrolysis to drive transport, and the secondary-active transporters that harness the energy of electrochemical gradients. Primary-active transporters involved in multidrug efflux belong mostly to the ABC (ATP-binding cassette) superfamily, and the secondary-active transporters belong to one of five protein groups, namely the ‘major facilitator’ (MFS), ‘small multidrug resistance’ (SMR), ‘resistance/nodulation/cell division’ (RND), ‘multidrug and toxic compounds extrusion’ (MATE), and ‘proteobacterial antimicrobial compound efflux’ (PACE) families.
The cell envelope of Gram-negative bacteria, which is a formidable protective barrier against environmental hazards including toxic compounds, comprises two membrane bilayers of distinct lipid composition and an interstitial periplasm with a mechanically reinforcing peptidoglycan matrix. Moving molecules through this barrier, whether it is the uptake of nutrients, the expelling of toxic compounds, or the transport of large effector proteins such as virulence factors, requires specialized machinery to negotiate a course through the different layers. Cytotoxic compounds, including clinical antibiotics, can be driven from the cell by intricate nanomachines that are energy-transducing tripartite assemblies spanning the envelope. Such machines generally include an outer membrane porin-like protein, such as TolC, a membrane fusion protein, such as AcrA, MexA or EmrA, and an inner-membrane protein, usually a transporter of the MFS, RND or ABC families. These pumps endow the bacteria with the capacity to tolerate a wide range of toxic compounds.
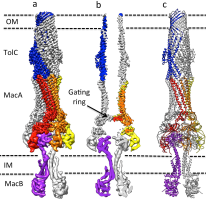
Fig 2. The ABC-type tripartite multidrug efflux pump MacAB-TolC at 3.3 angstrom resolution
The tripartite assembly AcrAB-TolC of Escherichia coli is a model system of proton motive force-dependent tripartite pumps. Crystal structures of the individual components of the pump have provided insight into how the proteins might function within the assembly. The outer-membrane component TolC forms a homotrimer with a porin-like transmembrane domain and a chute-like channel generated by a perisplasmic helical domain. The membrane fusion proteins such as AcrA and relatives are multi-domains structures with a hairpin architecture, and the inner-membrane proteins AcrB and other RND proteins form homotrimers with extensive periplasmic regions. Structural and mechanistic evidence shows that, in the presence of drugs, the AcrB trimer cycles through three structural states in a peristaltic pumping mechanism. To understand how the transport process operates, structural information is required for the organization and interaction of the subunits within a complete tripartite assembly. Guided by the crystal structures of the protomers, we have pioneered an approach of engineering components of the assemblies so that they form robust and functional complexes in vivo that withstand chromatographic purification and yield material of sufficient quality and quantity to permit structural analysis. Analyzing these engineered specimens by electron cryo microscopy, we have obtained the first experimental structure for a complete tripartite efflux pump (Du et al., Nature 2014). The current resolution of our reconstruction is approximately 3.6 Å, enabling crystal structures of the isolated components to be fitted and revealing a bound inhibitor (Wang et al., ELife 2017) (Fig. 1).
Applying the same engineering approach that has proven successful for the AcrAB-TolC pump, we have been able to generate stable assemblies of the ABC-driven MacAB-TolC pump. This fusion retains ATPase activity and fully supports drug efflux in in vivo assays. Our cryoEM reconstruction of the isolated MacAB-TolC complex is consistent with an assembly stoichiometry comprising one TolC trimer, six MacA monomers and one MacB dimer. TolC is held in an open state in this pump (Fitzpatrick et al. Nat Microbiol. 2017) (Fig. 2).
