The Hollfelder and Hyvönen labs have published a detailed molecular description of the chemistry at play during the evolution of a new function.
Promiscuous enzymes that accelerate multiple chemically distinct reactions were once regarded as oddities, as the enormous catalytic power of enzymes was thought to be irrevocably connected to precise substrate specificity. These 'sloppy' enzymes are now under scrutiny, as promiscuity offers a functional repertoire from which new chemistry can be recruited and further enhanced by mutations to facilitate adaptation. Therefore, the study of promiscuous enzymes not only broadens our understanding of molecular recognition, but also provides invaluable insights into how evolution alters these molecular interactions to achieve functional repurposing.
To investigate the emergence of a new enzymatic function from a promiscuous progenitor, Charlotte Miton, Stefanie Jonas and other members of the Hollfelder and Hyvönen labs performed experimental protein evolution in the test tube. Starting with an enzyme that predominantly hydrolysed sulfate esters, but which could also break down phosphate and phosphonate esters, they successfully increased phosphonate monoester hydrolysis by >100,000-fold, making this one of the biggest jumps in a directed evolution experiment. Remarkably, the large increase in the selected activity was accompanied by only modest changes in other promiscuous activities: the original activity (sulfatase) suffered a modest 400-fold decrease, while other promiscuous activities were marginally altered (30- to 53-fold changes). Instead of creating a specialist, evolution tailored a catalyst that can do more things efficiently! Good news for protein engineering, which is often challenging and frustrating, because new functions are difficult to create.
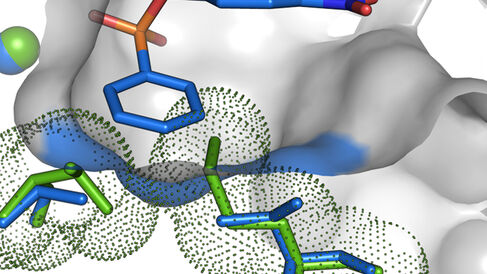
These observations raised the question of the changes happening at the molecular level: can we zoom into the catalytic cleft and understand how evolution directly altered the enzyme's chemistry? To this end, Miton and colleagues mapped the changes to the active site by generating structural snapshots of all evolved variants by X-ray crystallography, carried out kinetic analyses that defined constraints and degrees of freedom for catalysis, measured charge changes at the substrate/enzyme interface in the transition state (by linear free energy relationships), and confirmed possible binding orientation of the substrate in the active site by molecular dynamics simulations (in collaboration with Lynn Kamerlin's group at Uppsala).
Despite a 100,000-fold increase in phosphonate hydrolysis, the molecular picture that emerged suggests a subtle re-tuning of the molecular interactions between the promiscuous substrate and the evolved enzyme. At the structural level, two key mutations resolved a steric clash between the substrate and residues lining the active site cavity, unlocking the access to the chemical machinery for the phosphonate monoester. At the chemical level, the authors showed that by enabling the substrate to bind the enzyme in a more favourable orientation with respect to the catalytic centre, the negative charge building-up on the oxygen atom involved in bond breaking during catalysis (at the transition state) could be more efficiently neutralized by active site residues in its vicinity.
Thus, evolution created a new enzyme-substrate complex (ES, Michaelis) without the need for new catalytic residues to arise: rather than reinventing new catalytic features, the existing ones are deployed more efficaciously to act upon a reoriented substrate. These results suggest that such molecular tinkering may be a general 'recipe' for rapid functional transition in promiscuous enzymes: a high reactivity of permissive active site architectures allows multiple substrates to bind the enzyme in distinct orientations, as long as the chemical requirements for the reaction to occur are retained. The repurposing of chemical reactivity as a driver for evolution could constitute a 'simpler', more accessible solution for rapid adaptation and may be widespread. The intellectual framework employed to analyse this evolutionary trajectory will be of general use to understand how molecular evolution works in nature, and provide insights for mechanism-guided artificial evolution.
