The Taipale Group have published a new research article in the journal eLife.
DNA is the blueprint for life. It carries the genetic instructions required for all cells to produce proteins and some other molecules necessary for them to fulfil their functions, and for them to grow, develop and reproduce. These genetic instructions are contained within genes, regions of DNA that encode a particular function, with the information stored in the form of a code built from four different chemical bases (nucleotides); Adenine (A), Thymine (T), Cytosine (C) and Guanine (G). It is the order of these bases within a gene that determines the protein that gene will produce.
Although almost every single cell within an organism will contain the exact same genetic information, not every cell will require every protein, nor do they require all their proteins to be produced all of the time. For example, rhodopsin (the receptor protein involved in detecting light) is only produced in the retina, whilst cyclin B (a protein involved in cell cycle progression) is only required during cell division. To ensure the right genes are switched on to produce proteins in the correct cells and at the correct time, transcription factors (TFs) regulate gene activity.
TFs are a group of proteins that can glide along DNA and bind to short gene regulatory sequences to turn genes on or off. This recognition and binding of regulatory sequences can occur though three different mechanisms:
- Direct readout: specific bonds are formed between the TF and the chemical bases within the DNA.1-3
- Indirect readout: the TF recognises the DNA shape and electrostatic potential in the region of the regulatory sequence.4-8
- Water bridges: water molecules interact with both the TF and the chemical bases to link them together.9-15
These different mechanisms of binding will result in interactions of different strength between the TF and the gene regulatory sequence. To understand the interaction strength, we need to consider two physical concepts:
- Enthalpy, which concerns the energy involved. Chemical bonds formed directly between a TF and DNA in the direct readout mechanism give a stronger interaction than the water bridge mechanism where the TF and DNA would be linked indirectly via a water molecule.
- Entropy, a measure of disorder, where more disorder creates stronger binding. A water bridge creates more order than the direct readout mechanism as water molecules become involved in the interaction and can no longer move around freely. This results in a water bridge being a weaker interaction.
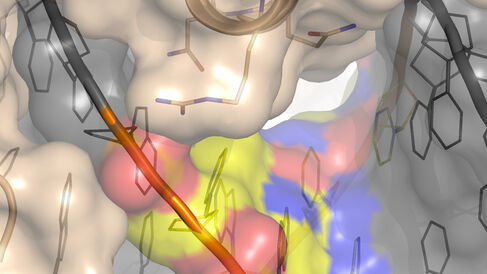
Most TFs can only bind gene regulatory sequences that are highly similar to each other. There are some TFs, however, that are more promiscuous and can bind several different sequences, although the mechanisms permitting this have not previously been clear.16-21 In their new publication, Morgunova et al. used X-ray crystallography, computational modelling and thermodynamic measurements to study four human TFs that can each bind to two different gene regulatory sequences. The authors found that the TFs bound each sequence with a similar strength, but through different mechanisms. Binding to one DNA sequence utilised an enthalpy-based mechanism where rigid water bridges linked the TF to the DNA. In contrast, binding to the second DNA sequence employed an entropy-based mechanism where the movement of water molecules increased disorder, giving strength to the interaction.
Looking ahead, a better understanding of the mechanisms through which TFs bind to DNA may aid efforts in developing new treatments that are dependent on the ability of molecules to bind one another. Moreover, the enthalpy/entropy-based binding mechanisms observed in this publication are likely to apply to many other molecules that interact through water bridges. In the future, these findings may help scientists predict the strength of an interaction solely through knowing the structures of the interacting molecules.
1Aggarwal et al., Science 242:899 (1988).
2Andrerson et al., Nature326:846 (1987).
3Wolberger et al., Nature335:789 (1988).
4Dror et al., Nucleic Acids Res.42:430 (2014).
5Hizver et al., PNAS98:8490 (2001).
6Joshi et al., Cell131:530 (2007).
7Lavery, Q. Rev. Biophys.38:339 (2005).
8Rohs et al., Structure13:1499 (2005).
9Bastidas and Showalter, J. Mol. Bio.425:3360 (2013).
10Garner and Rau, EMBO J. 14:1257 (1995).
11Ladbury et al., J. Mol. Bio.238:669 (1994).
12Morton and Ladbury, Protein Sci.5:2115 (1996).
13Patikoglou and Burley, Annu. Rev. Bioph. Biom.26:289 (1997).
14Poon, J. Bio. Chem.287:18297 (2012).
15Spolar and Record, Science263:777 (1994).
16Badis, Science324:1720 (2009).
17Johnson, Science316:1497 (2007).
18Jolma et al., Cell152:327 (2013).
19Morris et al., Nat. Biotech.29:483 (2011).
20Zhao and Stormo, Nat. Biotech.29:480 (2011).
21Zuo et al., Science Advances3:eaao1799 (2017).
