An article published in Nature Biotechnology by Katrin Fischer et al. from Florian Hollfelder’s group in collaboration with the groups of James Thaventhiran, Marko Hyvonen, Nick Matheson and Charlotte Deane establishes a new, generalised technology for finding potentially therapeutic antibodies in response to infectious disease outbreaks faster than currently possible, using anti-covid reagents as a showcase.
Covid was an example for a disease outbreak with enormous consequences and similar health challenges will become more likely in the future. Monoclonal antibodies are the most successful biologic drug modality and have proved effective for treatment and prevention of infectious diseases, such as COVID-19. They are especially important in the early stages of a viral outbreak when effective vaccines are not available yet, or in immunocompromised patient groups, where vaccination is ineffective. Once a suitable antibody therapeutic has been isolated, the race against escape mutants continues, potentially demanding routine updates of antiviral antibodies. In a pandemic it is therefore imperative that monoclonal antibodies can be generated rapidly. This accelerated demand – with timescales measured in months rather than the years of conventional pharmaceutical pipelines – required a new technology that enables faster responses, while remaining economical, easy to use, and available wherever specific viral challenges emerge.
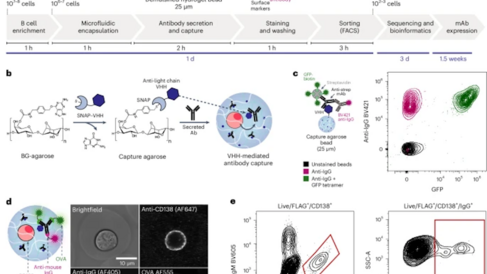
The article introduces a straightforward, ultrahigh-throughput technology to generate recombinant monoclonal antibodies from single antibody secreting cells, harvested from the human immune system in less than two weeks. Highly potent and broadly protective binders are rare, so the ultrahigh screening capacity increases the likelihood of isolating powerful drug candidates on a short timescale. Key to the technology is the combination of microfluidics and flow cytometry, enabling multi-parameter, ultrahigh throughput cell sorting. It illustrates its potential in pandemic preparedness by efficiently mining hundreds of new SARS-CoV-2-specific antibodies in one experiment.
This technology breaks new ground because:
(1) It facilitates the study of function-specific antibody secreting cells:Antibody secreting cells such as plasma cells are responsible for the active humoral response, producing antibodies for serological protection. So far, they have been understudied due to technical difficulties: most secreting cells lack surface B cell receptors and it is therefore challenging to map phenotype back to genotype. The technology solves this problem, enabling the rapid characterisation and sequencing of monoclonal antibodies from this underexplored immune compartment.
(2) It is high-throughput, fast, and low cost. The technology is simple, quick, and cheap to set up and run; only a standard microfluidic setup and commercial FACS machine are needed. Screening rates of >107 cells per day are routinely achievable, enabling the isolation of very rare cell populations and the comprehensive characterisation of the antigen-specific antibody response.
(3) It can be used to discover antibodies with clinical-grade activity. It isolates post-vaccination antibodies from a single sample with exceptionally high binding affinities (<10-12 M) and neutralising capacities (10 – 100 ng/mL) – comparable to the Regeneron clinical-stage antibody cocktail: instead of an industrial effort at extremely high cost, the antibodies are generated by a single researcher in two weeks with cheap reagents.
(4) It has a very high success rate. The discovery campaigns were highly efficient with a true positive rate of 95% for human antibodies. The technology saves time and money by accelerating the route to successful antibodies.
(5) It is versatile and can be readily adapted to specific use cases. The scope of the technology is not limited to human antibodies or single antigens: the modular approach enables interrogation of cells from different species and screening of several different antigens or variants in a single experiment. Therefore, the technology can be used to isolate antibodies with specific binding modes or as an alternative to hybridoma technology to meet the large demand for well-defined monoclonal antibodies in research and diagnostics.
The project would not have been possible without a broad interdisciplinary collaborative effort spanning skills from clinical work and immunology to structural biology, organic chemistry and microfluidic engineering, all centred around the global challenge of pandemic preparedness. The groups involved (James Thaventhiran, Marko Hyvonen, Nick Matheson and Florian Hollfelder plus an Oxford collaborator, Charlotte Deane) are from different departments (Medicine, Biochemistry, MRC Toxicology, Computational Biology), underlining the collection of expertise that was necessary to crack this problem. The work is building on Cambridge’s research priorities, including the strategic research initiative ‘Precision Health’, the strategic research network ‘Immunology’ and is in the remit of the interdisciplinary centres ‘Engineering Biology’, the ‘Cambridge Centre for Data-driven Discovery’ and the ‘Cambridge Academy of Therapeutic Sciences’.
