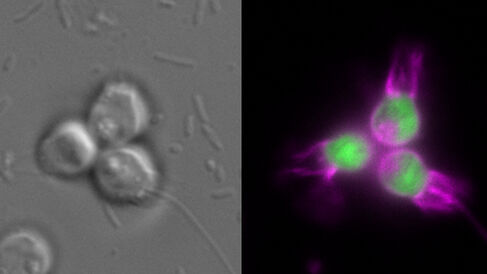
Dr Ross Waller, and collaborators from across the globe, have published a new paper in PLoS Biology describing a new community-based approach for developing model marine protists.
Biological research frequently involves the use of model organisms for the study of particular processes or traits, with the expectation that the discoveries made will provide insight into the workings of other organisms. Often, these models are chosen as their specific characteristics makes them more amenable to experimental studies; characteristics such as a short life cycle, available techniques for genetic manipulation, and non-specialist living requirements.
In eukaryotes, yeasts such as Saccharomyces cerevisiae have been widely used for cell biology research as they are quick and easy to grow, and as they have a similar cell cycle to humans that is regulated by homologous proteins. Drosophila melanogaster has been exploited because of its short life cycle, and because of a relatively simple method being available to disrupt its genes via transposable elements for genetic studies. The roundworm Caenorhabditis elegans is also commonly studied and is critical for developmental research due to its highly defined development patterns involving fixed numbers of cells. Moreover, model organisms are essential for human disease research where human experimentation would be unethical.
Research utilising model organisms has unequivocally enabled tremendous progress in our understanding of life. However, the small number of available model organisms, and the fact that the majority of these are all drawn from the animal branch of the tree of life, gives a restricted view of biological diversity where just a few relatively closely related organisms, in evolutionary terms, have been used to define 'the canonical' for all eukaryotes.1-2
Eukaryotes are, in fact, far more diverse than is appreciated by our current complement of model organisms. Much of this diversity is represented by the single-celled protists, which are more divergent from one another than animals are from fungi.3 This is reflected in the striking differences in genetic content, forms, behaviours, and ecological function found across the protozoan kingdom. Understanding the full breadth of protozoan diversity is essential for understanding ancestral eukaryotic characteristics and diversification, as well as for understanding the acquisition and evolution of organelles and the origins and diversity of processes as fundamental as sex.4-7 Unfortunately, the protozoan lineage lacks experimental models, and marine protists in particular suffer from a paucity of model organisms despite them playing critical roles in global nutrient cycles, food webs, and climate.8
Many obstacles and uncertainties need to be faced in developing new model marine organisms: Can DNA or RNA be introduced into the organisms? Will the genes be expressed? Can transformants be selected? Will the organisms tolerate these treatments? In their new publication, Waller et al. describe a new community-based approach to address these challenges that was developed through the Gordon and Betty Moore Foundation's Marine Microbiology Initiative: Experimental Model Systems (EMS) Programme. Here, 34 project teams were selected to develop new genetic tools for marine protists, with nine focusing on protists for which basic genetic tools were already available and the remainder investigating an array of protists where reliable transformation had yet to be achieved. The EMS goal was that scientists studying the lesser explored organisms would learn directly from the successes made with the more advanced model protists through strong community building, open dialogue between research groups, and enhanced data sharing (including that of negative results). Over the course of two years, the EMS Programme has created a novel model for research that has enabled substantial progress in the development of genetic tools for marine protists, with stable transformation achieved in six major protist taxonomic classes and transient transgene expression achieved in a further six classes.
1Goldstein and King, Trends Cell Biol. 26:818 (2016).
2Russell et al., BMC Biol. 15:55 (2017).
3Burki et al., Proc. Biol. Sci. 283 (2016).
4Carradec et al., Nat. Commun.9:373 (2018).
5Archibald, Curr. Biol. 25:R911 (2015).
6Loidl, Annu. Rev. Genet.50:293 (2016).
7Speijer et al., PNAS112:8827 (2015).
8Worden et al., PLoS Biol. 12:e1001889 (2014).
