Characterisation of the modulation of host cell RNA-binding proteins through the infectious cycle of SARS-CoV-2 and the interactions that the viral RNA establishes with the host proteins.
Kathryn Lilley, along with her long-term collaborator, Anne Willis (MRC Toxicology Unit), have formed a consortium of researchers in Cambridge, Oxford, Leicester and Munich in a very short period of time, along with collaborators from the pharmaceutical industry, to interrogate host protein-viral RNA interactions.
The SARS-CoV-2 virus responsible for the COVID-19 pandemic is an RNA virus, which means that, unlike in the case of humans, its genetic material is comprised of RNA. Once inside a cell, the viral RNA must be converted or 'translated' into proteins which then bring about its replication and evasion of the cell's defence mechanisms. Ultimately more viral RNA needs to be made and packaged into new virus particles inside the host cell and then released to go on to infect other cells. To enable all of the above, the virus has to make use of proteins that already exist in human cells. We aim to determine which human proteins are commandeered by the virus during its infection cycle and are necessary to enable it to reproduce itself. Our hope is that there are drugs that already exist that could disrupt the interactions between the viral RNA and host proteins.
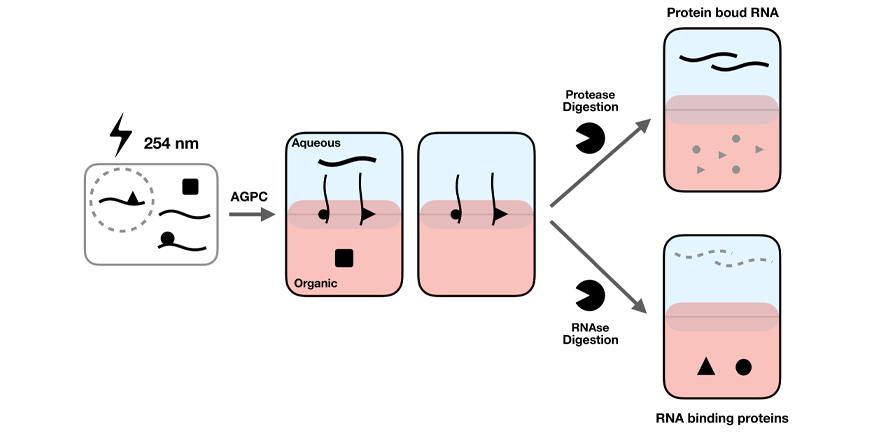
The Lilley and Willis Groups have developed an efficient, reproducible method (OOPS) to determine the RNA-binding proteome of cells. We are applying OOPS to upper respiratory tract-derived (normal and infected) organoids from Joo Hyeong Lee's Group at the Stem Cell Institute in Cambridge, to determine the manipulation of the RNA-binding proteome upon viral infection of the organoids over an infection cycle.
OOPS is also being applied to Calu-3 (a human lung epithelial cancer cell line) in order to determine modulation of the host RNA-binding proteome upon infection and direct interaction of RNA-binding proteins with the viral transcriptome. This work will allow identification of the host-viral RNA-protein interactions and enable an understanding of how the cell responds to the virus, and how the virus manipulates the cellular RNA-interacting machinery and, in particular, the translation apparatus.
Altogether, this information will be used to map the RNA interactions onto protein structure and identify drugs that are already clinically available as possible treatment options.
In a separate project, the Lilley Group is collaborating with Markus Ralser (Charité Medical University of Berlin and the Francis Crick Institute) to establish plasma protein markers of the severity of COVID-19 in patients. Here they are applying algorithms developed by Vadim Demichev (DIA-NN) to very-high-throughput mass spectrometry analysis of plasma proteins.
Publications
Messner CB, Demichev V, Wendisch D, Michalick L, White M, Freiwald A, Textoris-Taube K, Vernardis SI, Egger A-S, Kreidl M, Ludwig D, Kilian C, Agostini F, Zelezniak A, Thibeault C, Pfeiffer M, Hippenstiel S, Hocke A, von Kalle C, Campbell A, Hayward C, Porteous DJ, Marioni RE, Langenberg C, Lilley KS, Kuebler WM, Mülleder M, Drosten C, Suttorp N, Witzenrath M, Kurth F, Sander LE, Ralser M (2020). Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst., S2405-4712(20)30197-6. doi: 10.1016/j.cels.2020.05.012
Figure from: Queiroz RML, Smith T, Villanueva E, Marti-Solano M, Monti M, Pizzinga M, Mirea D-M, Ramakrishna M, Harvey RF, Dezi V, Thomas GH, Willis AE, Lilley KS (2019). Comprehensive identification of RNA-protein interactions in any organism using Orthogonal Organic Phase Separation (OOPS). Nat. Biotechnol., 37(2):169-178. doi: 10.1038/s41587-018-0001-2
