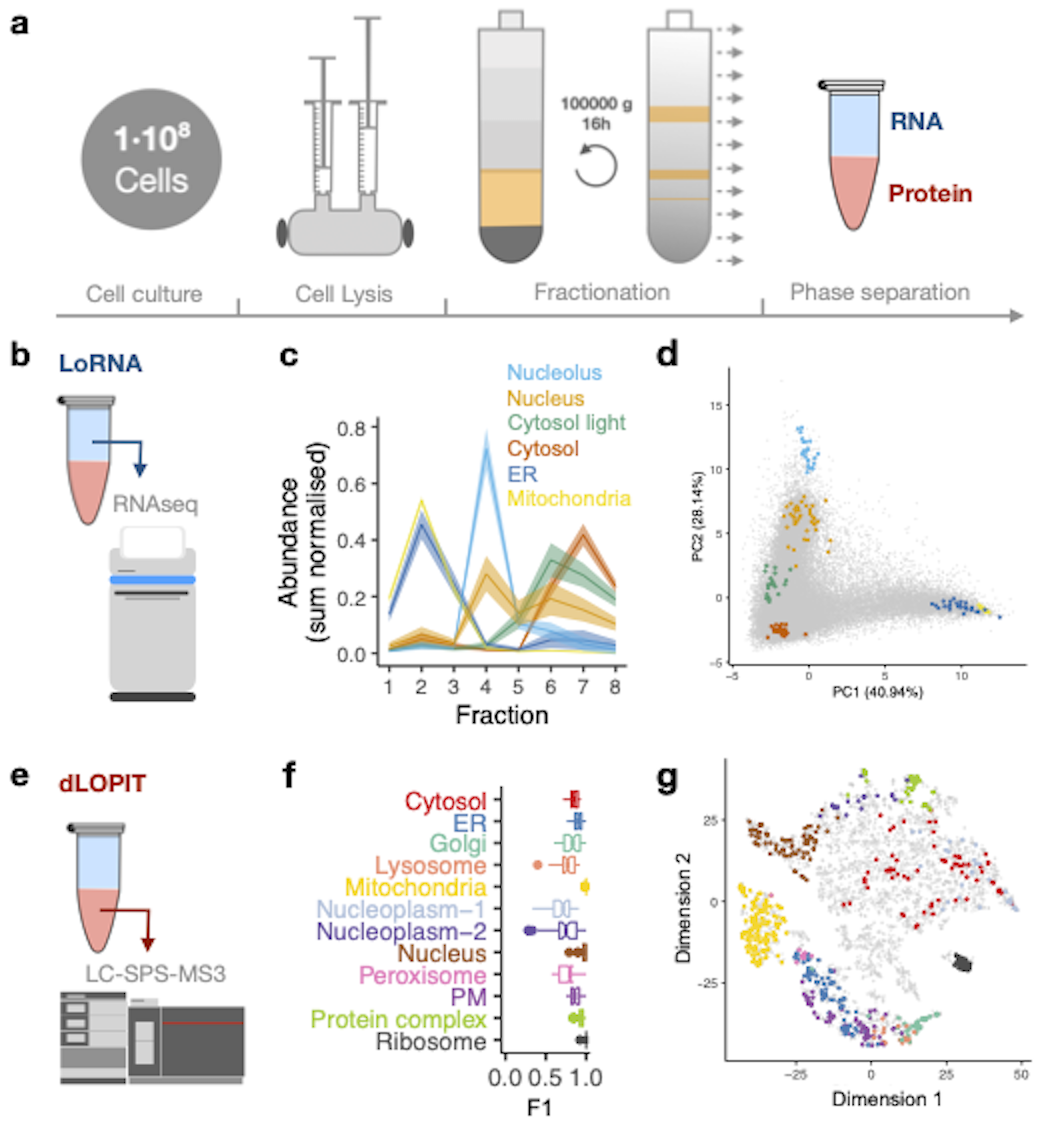
A paper published in Nature Methods presents the most comprehensive overview to date of RNA and protein subcellular localisation dynamics.
The research groups of Kathryn Lilley in the Biochemistry Department, Cambridge and Anne Willis from the MRC Toxicology Unit together with colleagues in the Structural Biology Research Centre, Milan and the Department of Statistics, Oxford, present an integrative framework to simultaneously interrogate the dynamics of the transcriptome and proteome at subcellular resolution.
They have extended the subcellular proteomics approach developed in the Lilley group, Localisation of Proteins by Isotope Tagging (LOPIT), to co-capture the Localisation of RNA (LoRNA) to -based allow integration all RNA and protein subcellular locations in a single experiment. LoRNA enables system-wide quantification of the proportional distribution of RNA. Using this approach, they have revealed that ER-localised transcripts are more efficiently recruited to cytosolic granules than cytosolic RNAs, and that the translation initiation factor eIF3d is key to sustaining cytoskeletal function by obtaining a cell-wide overview of localisation dynamics for 31839 transcripts and 5314 proteins during the unfolded protein response (UPR).
Professor Lilley said:
‘One of our most striking findings is that membrane-localised mRNAs are recruited more efficiently to granules than cytosolic mRNAs, while lncRNAs do not relocalise to granules. We show that the two features most associated with mRNA recruitment to granules, length and AU content do not affect lncRNA relocalisation to granules. This suggests that RNA recruitment to granules may depend on more than RNA length and ribosome occupancy or that there is a mechanism to exclude lncRNAs from granules.’
By simultaneously characterising proteome and transcriptome relocalisation, we can transform the study of how the cell co-ordinately responds to physiological signals, stress conditions, exogenous cues, or infectious pathogens. The approach will contribute to the translation of molecular biology observations to medical benefits by fostering new studies into the role of RNA and protein localisation dynamics in cell homeostasis and disease.
